-
Notifications
You must be signed in to change notification settings - Fork 1
/
Copy path03-data-manipulation-examples.Rmd
284 lines (188 loc) · 6.99 KB
/
03-data-manipulation-examples.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
---
title: "Data Manipulation Examples"
author: "Joscelin Rocha Hidalgo"
output:
html_document:
css: slides/style.css
toc: true
toc_depth: 1
toc_float: true
df_print: paged
---
```{r setup, include=FALSE}
knitr::opts_chunk$set(echo = TRUE)
```
# Loading Packages
Load the `tidyverse` package.
```{r}
library(tidyverse)
```
# Import chds6162_data
```{r}
data <- read_csv("data/chds6162_data.csv")
```
# select

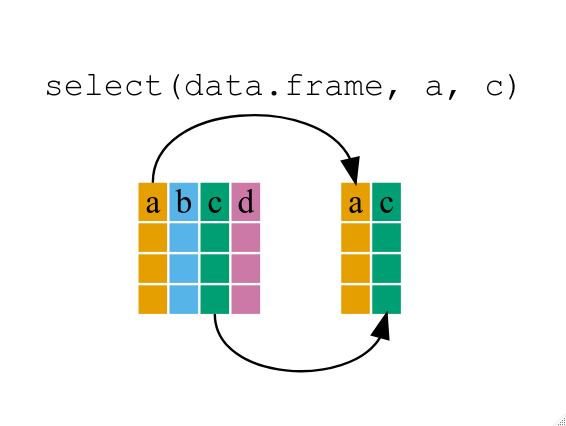
With the function `select` we can select variables (columns) from the larger data frame.
Use `select` to show just the `gestation` variable.
```{r}
data_ges <- data %>%
select(gestation)
```
We can also `select` a range of columns. `select` all the variables that belong to the father (they had a "d" in front of them) `drace` to `dwt`.
```{r}
data %>%
select(drace:dwt)
#What about just the id column and everything after the father information?
data %>%
select(id, marital:last_col())
```
We can drop variables using the -var format. Drop the `marital` variable.
```{r}
data %>%
select(-(marital))
```
# mutate


We use `mutate` we make new variables or change existing ones.
Create a **new variable with a specific value**
Create a new variable called `data_decade`. Imagine that you will be merging this dataset from 61-62 to dataset from the 70's. To make it easier, you will create this variable with the value "60s."
```{r}
data %>%
mutate(data_decade = "60s")
```
Create a **new variable based on other variables**
Create a new variable called `wt_k`. This variable will give you information about mom's weight pre-pregnancy(`wt`) in kilos (1 pound = .454 kilos).
```{r}
data %>%
mutate(wt_k = wt*.454)
# too many decimals? let's round things
data %>%
mutate(wt_k = round((wt*.454),2))
```
# case_when

Change an **existing variable**
For some reason you want to have the real labels rather than the values. Let's change the `marital`variable to its real values: 1 = married, 2 = legally separated, 3 = divorced, 4 = widowed, 5 = never married.
```{r}
data %>%
mutate(marital = case_when(
marital == 1 ~ "married",
marital == 2 ~ "legally separated",
marital == 3 ~ "divorced",
marital == 4 ~ "widowed",
marital == 5 ~ "never married"
))
```
# filter

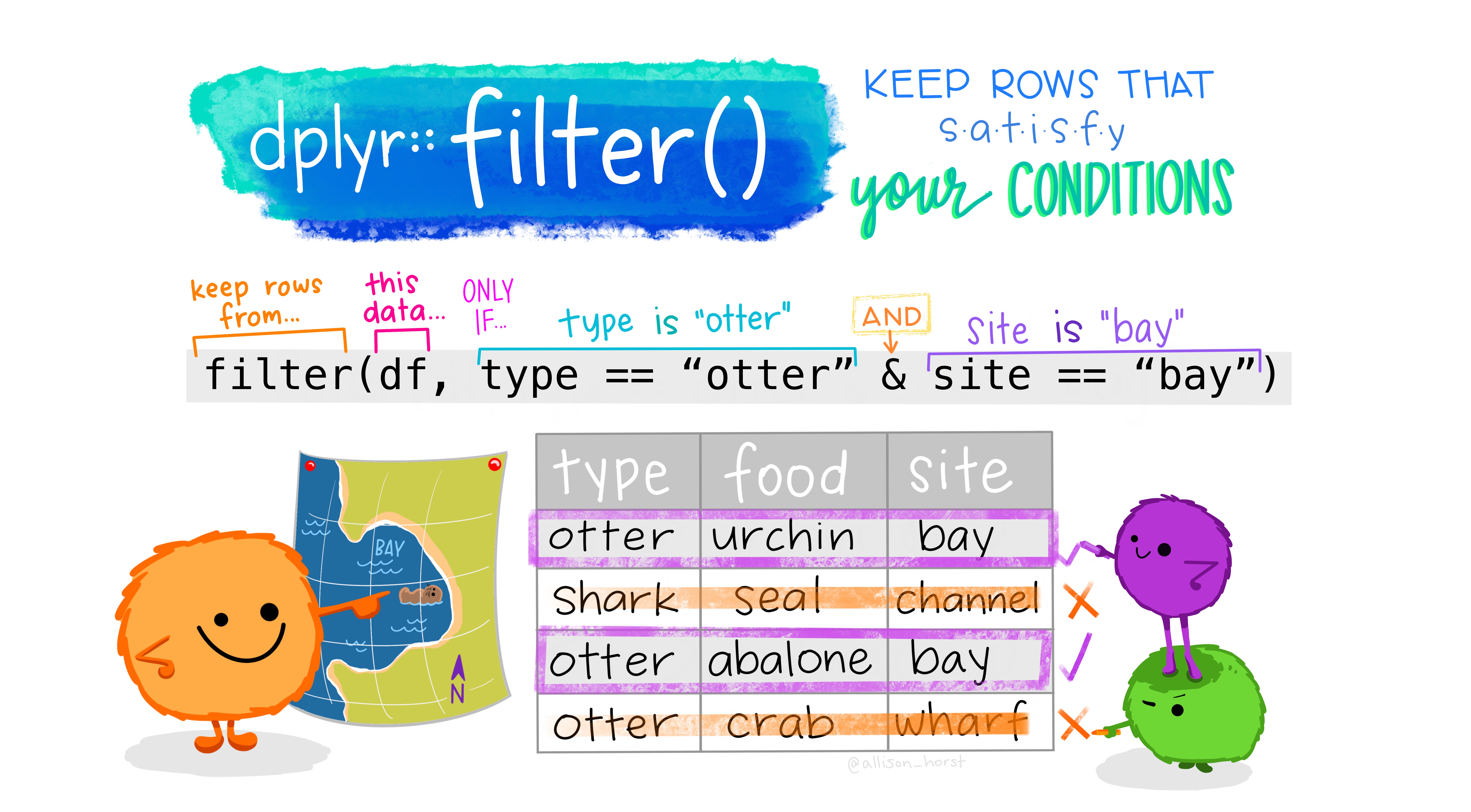
art by @allison_horst
We use `filter` to choose a subset of cases.
Use `filter` to keep only those whose parents have been never married (value `5` in the `marital` variable. Then, use `select` to show only the `age` variable to let us explore the mothers' ages when they delivered.
```{r}
data %>%
filter(marital == 5) %>%
select(age)
```
Use `filter` to keep only those who are **not** divorced (value `3`).
```{r}
data %>%
filter(marital != 3)
```
Use `%in%` within the `filter` function to keep only those who are divorced (3), legally separated (2), or widowed (4).
```{r}
data %>%
filter(marital %in% c(2, 3,4))
```
Create a chain that keeps only those whose moms were in their 20s when they delivered and are college grads (val= 5). Use the `age` and `ed` variables.
```{r}
data %>%
filter(ed == 5, age %in% 20:30)
```
We can use `<`, `>`, `<=`, and `>=` for numeric data. == equal, != not equal
# summarize or summarise

With `summarize`,as the name implies, you will get a summary of your dataset.
Get the mean days of gestation for this sample.
```{r}
data %>%
summarize(gestation_length_mean = round(mean(gestation, na.rm = TRUE),2))
```
We can have multiple arguments in each usage of `summarize`.
In addition to the mean gestation length you calculated earlier, calculate the minimum and maximum for the same variable.
```{r}
data %>%
summarize(mean_gestation_length = mean(gestation, na.rm = TRUE),
min_gestation_length = min(gestation, na.rm = TRUE),
max_gestation_length = max(gestation, na.rm = TRUE))
```
# group_by


`group_by`enables us to perform calculations on each of our groups.
Calculate the mean gestation length in days based on mother's smoking habits (0=never, 1=smokes now, 2=until current pregnancy, 3=once did, not now) using `group_by` and `summarize`.
```{r}
data %>%
group_by(smoke) %>%
summarize(mean_gestation_length = mean(gestation,
na.rm = TRUE))
#Since we know that there are some NAs in the Smoke variable we can go ahead and drop those NAs before we summarize them
data %>%
drop_na(smoke) %>%
group_by(smoke,ed) %>%
summarize(mean_gestation_length = mean(gestation,
na.rm = TRUE))
```
We can use `group_by` with multiple groups.
# across

`across`enables us to perform calculations on each of our selected columns
In the example above, we calculated the mean for gestation but what about if I wanted the mean for gestation, mom age (age) and dad age (dage)
```{r}
#If we didn't know better, we would do this:
data %>%
drop_na(smoke) %>%
group_by(smoke) %>%
summarize(mean_gestation_length = mean(gestation,
na.rm = TRUE),
mean_m_age = mean(age,
na.rm = TRUE),
mean_d_age = mean(dage,
na.rm = TRUE))
```
```{r}
#But we know better:
data %>%
drop_na(smoke) %>%
group_by(smoke) %>%
summarize(across(c(gestation,age,dage),mean,na.rm = TRUE))
?across
```
# Create a new data frame
Sometimes you want to save the results of your work to a new data frame or replace the one you already have (not recommended).
```{r}
to_graph <- data %>%
drop_na(smoke) %>%
group_by(smoke) %>%
summarize(across(c(gestation,age,dage),mean,na.rm = TRUE))
```
# relocate
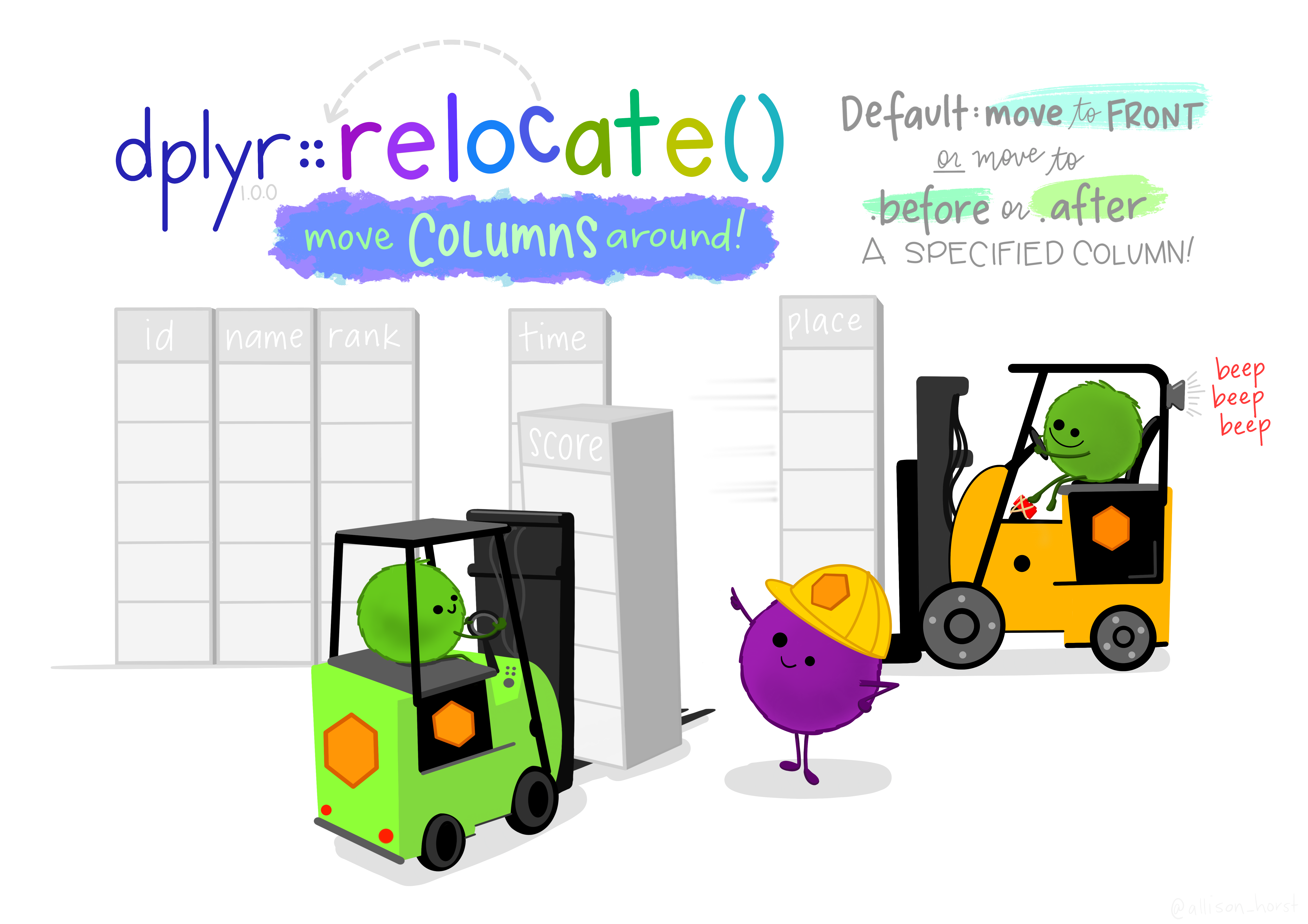
Sometimes you want to export the table as a csv but you don't really like the order of the columns. Relocate allows you to move them around without you having to go back to previous codes. Let's take the `to_graph` data frame and move the age column before the gestation column.
```{r}
?relocate
to_graph %>%
relocate (age, .before = gestation)
#What if I wanted gestation to be the last column?
to_graph %>%
relocate (gestation, .after = last_col())
# ok now I like it and want a CSV of it.
# I make sure to save it as a dataframe and then export it
csv_to_export <- to_graph %>%
relocate (gestation, .after = last_col())
# export csv
write_csv(csv_to_export, "exports/example_to_export.csv")
```