The two primary goals of the OTree algorithm are
- enable spatial datasets with multi-dimensional indexing and
- empower the query engine with optimized sampling operators, that is, the ability to retrieve a statistically significant fraction of the dataset without reading all the data.
One of the most important techniques used to build a multi-dimensional index is through recursive space division; a bounded vector space initially containing all the data is recursively divided into equal-sized, non-overlapping subspaces, as long as they exceed the predefined capacity.
For a dataset indexed with n columns, the constructed index is an n-dimensional vector space composed of cubes, with non-overlapping boundaries. Each cube can contain a predefined number of element cap, and exceeding it would trigger recursively dividing a cube into child cubes by halving the ranges in all dimensions until the number of elements included no longer exceeds cap.
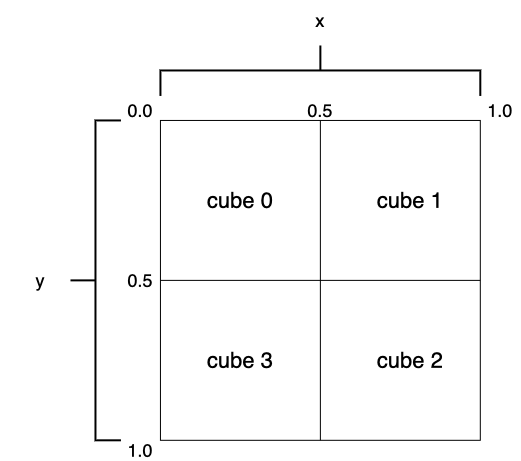
Say that we use two columns, x, and y to build the index, and the parameter cap for each cube is 2. The first image in the figure below is the root cube, containing more than two elements. The cube is split into four equal-sized, non-overlapping child cubes with one space division step, as shown in the middle image. Three of the four cubes are in good condition as a result of the division.
The process doesn't end here. The algorithm would keep splitting the space for the top-left cube until meeting the condition size ≤ cap for all cubes. In this particular case, it took four more divisions to finish. The third image shows the final result.
The figure below shows a treelike representation of the above example. Each square represents a cube, with green cubes that satisfy the requirement size ≤ cap and red ones that don't.
As stated earlier, all cubes have bounded values for the indexed columns, and they dictate the elements they can store. If we were to query a dataset and select a set of elements that fall inside the boundaries of cube 020, then all subsequent cubes 0200, 0201, 0203, 02020, 02021, 02022, and 02023 are to be consulted and read.
It is worth mentioning that each time we divide a cube, the cube itself would no longer hold the data but have it distributed among its child cubes. We store no actual data but their pointers in the index.
A sample operator, present in many query engines, can retrieve a randomly generated subset of data from the original dataset. In the era of massive datasets, the I/O involved in reading the entire TB or PB of data is both computationally expensive and time-consuming. Fortunately, one does not need all the data for many analytical workloads, and a statistically representative subset usually can do the trick.
However, although the result of a sample operation is a subset of the desired size, the price for generating such a subset is generally not lower - we first load the entire dataset into memory, and only then a random selection is used for the subset generation.
Built on top of a recursive space division algorithm, the OTree "Index" adopts a data denormalization methodology to empower both multi-dimensional indexing and efficient data sampling at the same time. It arranges the data and its replicas in a multi-dimensional tree, with no need for a separate structure for the index, for the architecture of the data arrangement itself already plays the role.
In a nutshell, we distribute our data and replicas among a multi-dimensional tree, where each cube gets to have a random subset of it. We can access this fraction through a parameter called maxWeight, and all cubes also have one of the three possible states that determine their READ and WRITE protocols.
In this case, all cubes store actual data and don't use pointers.
The rest of the page describes the theoretical details about the OTree, including cube states and their READ and WRITE protocols, how data replication is conducted.
-
cube: similar to a node in a tree data structure, except it defines a confined n-dimensional space, withnbeing the number of indexed columns. -
payload: elements stored in a cube that won't be cut off after replication -
offset: where the excess elements are stored for cubes withsize ≥ desired_capacity. -
desiredCubeSize: the capacity of thepayload. -
size: the number of total elements currently contained in a cube(payload + offset) -
weight: random integer value from [Int.MinValue, Int.MaxValue] assigned to new elements upon writing. Weights have a uniform distribution. -
maxWeight: the largestweightfrom all elements contained in a cube'spayload. They are the fraction of the dataset in each cube, and by design, are arranged in a non-decreasing order traversing the tree from top to bottom. -
maxElement: the element in the payload associated with themaxWeight. More than one element may have theweight = maxWeight, but only one can be themaxElement. -
indexedColumns: columns from the dataset used to construct the index
-
Sample fraction
ffromdf.sample(f)is the target sample size. A list of cubes that make up the fraction is constructed by analyzing the tree. -
READs start from the root and traverses the tree in a DFS fashion, prioritizing children over siblings.
-
The traversal ceases going downwards once a cube with
maxWeight > fis encountered. -
Sibling cubes from the current level are examined to determine their candidacy for the list.
-
whether a cube is to be included and what part of it should be read is determined by its state and the relationship between
fandmaxWeight -
The protocol ends by reading all cubes from the list.
-
Write an element
E(a, b | w)to the tree, with(a, b)being the values of the element for indexed columns(x, y), andwthe randomly assignedweight. -
From the root, find the proper cube among
cube 0,cube 1,cube 2, andcube 3forEaccording to its values(a, b). Say that columnsx, andyboth have the range [0.0, 1.0], and(a, b) = (0.1, 0.2). In this case,cube 0is the cube of choice.
-
Proceed to conduct the WRITE according to the WRITE protocol of the cube:
maxWeight > w: writeEaccording to the WRITE protocol of the cube dictated by its state. ThemaxWeightdefines the fraction of the dataset contained in the cube. Writing a new element to a full cube entails pushing themaxElementto the offset and update themaxWeight.maxWeight <= w: go to the correct child cube amongcube 00,cube 01,cube 02, andcube 03, and recheck the condition.
-
Unlike the traditional space division algorithm, we don't split the cube immediately when exceeding the capacity during writes. The optimization of the cube, and the index in general, is handled by a separate procedure.
The state of a cube dictates, among other things, its READ and WRITE protocol and whether replication of its contained elements exists among its subtree.
The following image depicts the three possible states, and whether a cube is of one state or another depends on the following factors:
size/desiredCubeSizeratio- the state of its ancestors
- whether
analyze()oroptimize()is called
-
FLOODED
This is the initial state of the cube. The protocol is separated into two cases.
-
size <= desiredCubeSize:maxWeightis set toInt.Maxvalue- READ: read elements from the
payloadwithweight <= f - WRITE: write to the
payload
- READ: read elements from the
-
size > desiredCubeSize:- READ:
f >= maxWeight: read thepayload + offsetf < maxWeight: read elements from thepayloadwithweight <= f
- WRITE:
- write to the
payload
- write to the
- READ:
-
-
ANNOUNCED
- WRITE: write to the
payload, mark the new elements asAfter Announcement(AA) - READ:
f >= maxWeight: read thepayload + offsetexcludingAAelementsf < maxWeight: read elements from thepayloadwithweight <= f
- WRITE: write to the
-
REPLICATED
The final state of any cubes where all elements are replicated and distributed among their subtrees.
- WRITE: write to
payloadand replicate the newly written element down the subtree - READ:
f >= maxWeight: don't read anythingf < maxWeight: read elements from thepayloadwithweight <= f
- WRITE: write to