R package of Drake-based tools for the Larman Lab's PhIP-Seq data analysis pipeline.
if(!requireNamespace("remotes")) install.packages("remotes")
remotes::install_github("ropensci/drake") # drake is on CRAN, but phipmake depends on the more recently updated Github version.
remotes::install_github("brandonsie/phipmake")drake::expose_imports(phipmake) # allow drake to keep track of nested function dependencies.
plan <- phipmake::define_plan() # initialize pipeline. looks for mpath.txt and ppath.txt in working directory
drake::make(plan) # execute pipeline
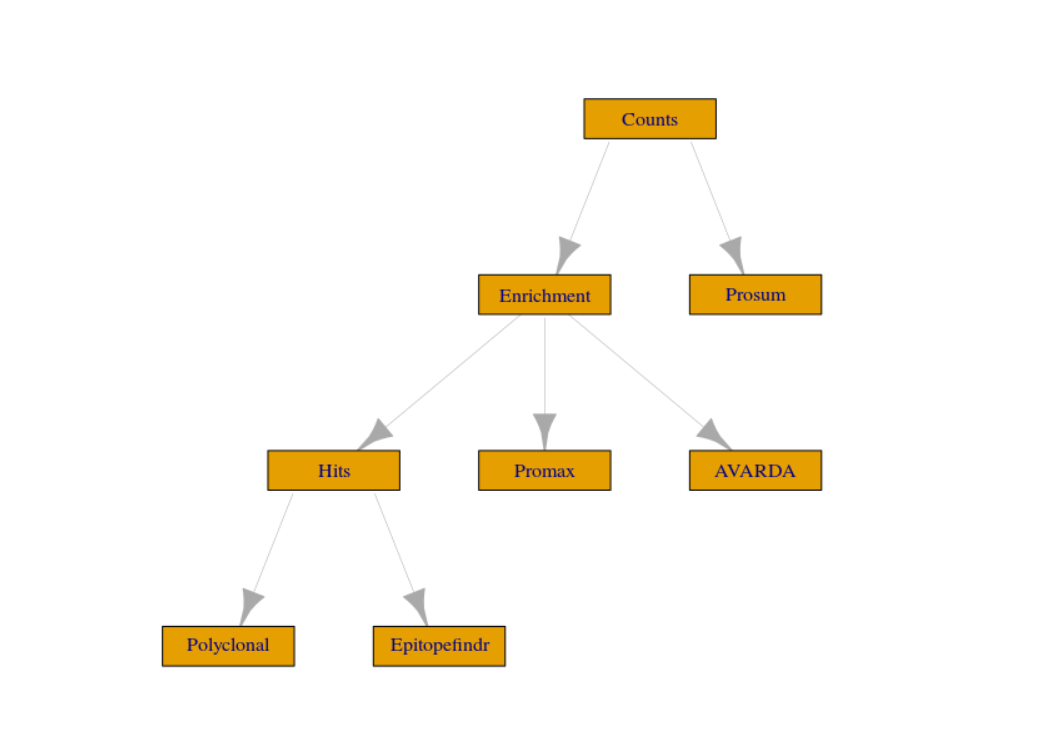
v <- drake::vis_drake_graph(drake::drake_config(plan)) # visualize pipeline dependencies
v
visNetwork::visSave(v, "dependency_graph.html") # save dependency graphA simplified version of the dependency graph is below. For a more complete & interactive representation, please click here.
More documentation will come soon to phipmake's website.
- Daniel Monaco wrote the network filter used for polyclonal scoring, and AVARDA, used for deconvoluting viral proteome antibody specificitity data.