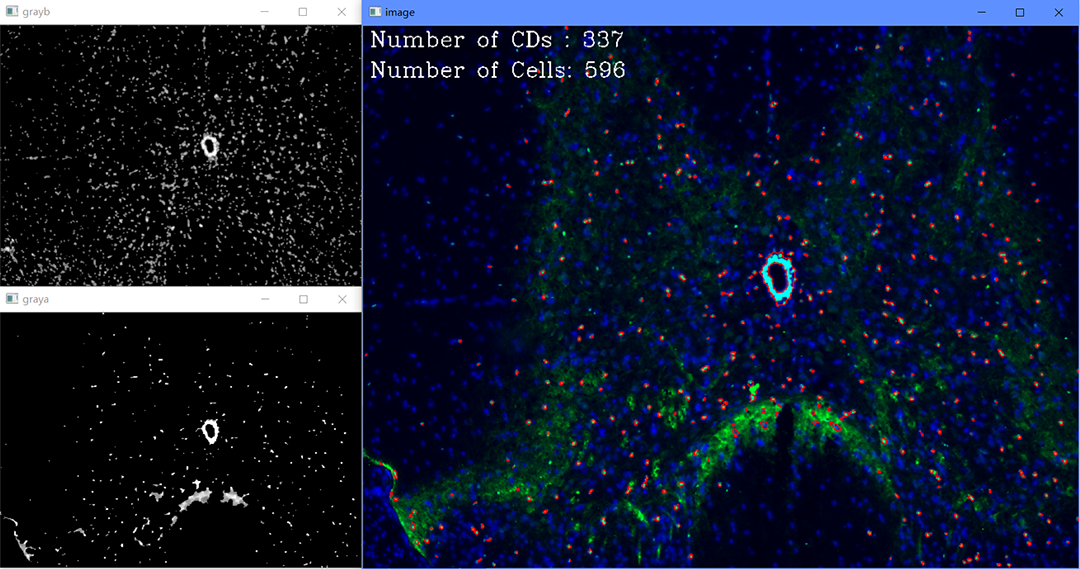
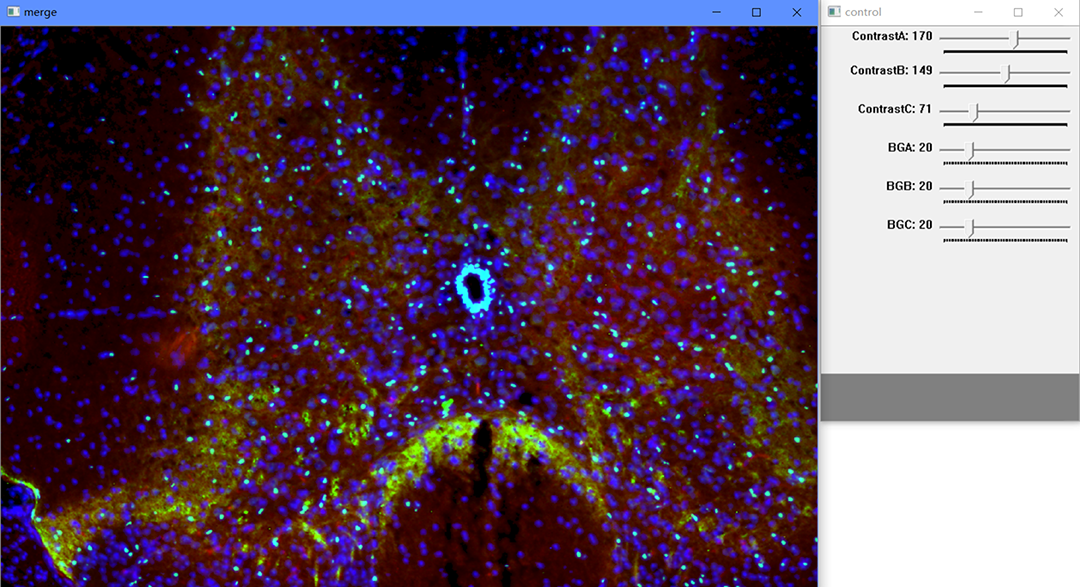
A series of tools to process the results of immunofluorescence in auxiliary.
windows
openCV with its related libraries
First you should rename your images by the following rule:
info_id.ext
-
info: the information of the image.
-
id: the sequence from 0 to 2.
*The image “_0” should be nuclear dyeing like DAPI.
-
ext: the extension of the image.
*Be sure that the format of the image is supported by openCV such as png, jpg, tif, etc.
# examples
br_0.tif
br_1.tif
For single image processing:
You can just drag the image "_0" to CellCount.
In addition, you can use the command line:
# single image processing.
CellCount info_0.ext
# example:
CellCount br_0.tif
For batch processing:
You can put the images in the folder "img" then run CellCount.
*The images to merge should have different main-channel such as R, G or B.
For single image processing:
You can just drag the image "_0" to CellMerge.
In addition, you can use the command line:
# single image processing.
CellMerge info_0.ext
# example:
CellMerge br_0.tif
You can adjust the parameters by dragging the track bars in "control" window.
Furthermore, you can move and scale the image by mouse in "merge" window.
- ContrastA: the contrast of the image "_0".
- ContrastB: the contrast of the image "_1".
- ContrastC: the contrast of the image "_2".
- BackGroundA: the limit of the intensity to reserve in the image "_0".
- BackGroundB: the limit of the intensity to reserve in the image "_1".
- BackGroundC: the limit of the intensity to reserve in the image "_2".
These parameters would be recorded in "cellMerge.ini"
For batch processing:
You can put the images in the folder "img" then run CellMerge.
The parameters for batch processing would be read from "cellMerge.ini"
The image "_m" is the visual result of CellMerge.
The image "_r" is the visual result of CellCount.
"result.tsv" is a Tab-separated table include:
- Type: the magnification of the image.
- CDs: the number of the nuclear areas.
- Cells: the number of the nuclei.
- M1CDs: the number of the merged areas about the image "_1".
- M1Cells: the number of the merged cells about the image "_1".
- M2CDs: the number of the merged areas about the image "_2".
- M2Cells: the number of the merged cells about the image "_2".
- MCDs: the number of the merged areas.
- MCells: the number of the merged cells.
- Info: the mean area of the nuclei.
v0.6a - Add 3 merged count
v0.5a - A new beginning
This code is BSD. Please check the LICENSE file for contributors.
The samples for testing are provided by Group of Wang SF, KMMU.
The released executables have been packed by upx. See https://upx.github.io for more details.