-
Notifications
You must be signed in to change notification settings - Fork 6
PanPhlAn applications and examples
Based on shotgun metagenomic samples, PanPhlAn enables:
- strain identification and characterization of unknown strains in metagenomic samples. The gene set of strains present in samples is detected by screening for all potential genes from the species pangenome.
- outbreak monitoring: pathogen detection and characterization, see E. coli example below.
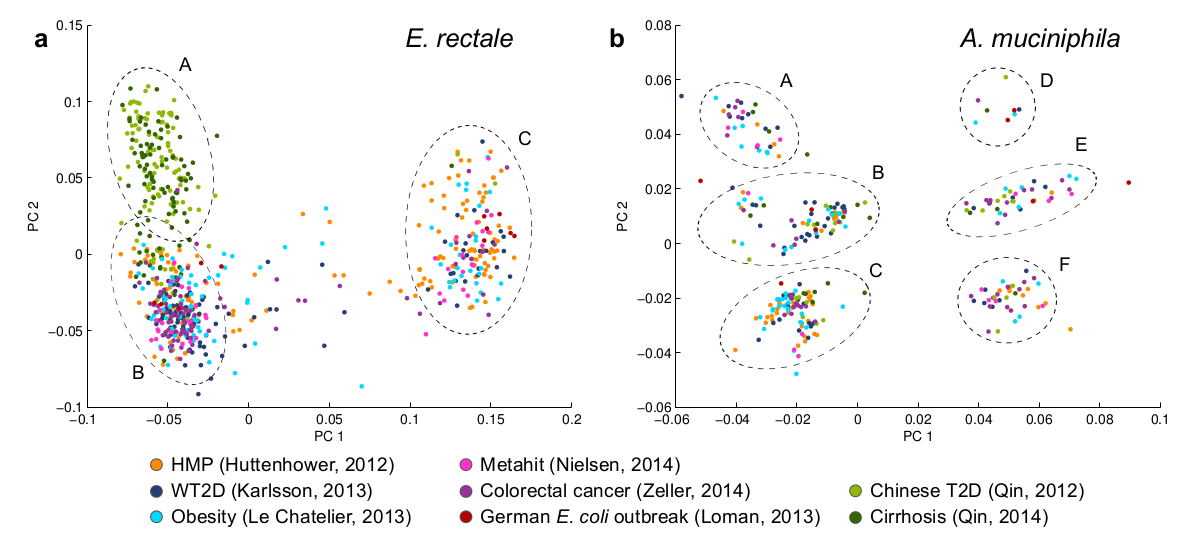
- population genomics: exploring the diversity of a species based on detected strains in hundreds of samples, see E. rectale and A. muciniphila examples below.
- strain tracking: detecting identical gene content profiles of strains in different samples
- functional analysis: based on detected strain-specific genes, the gene sequences can be used for functional investigations using KEGG or other similar databases.
PanPhlAn profiling of the German outbreak metagenomes using a reference database in which the target outbreak genome is missing. (a) Hierarchical clustering. The heatmap displays presence/absence gene-family profiles of 110 reference strains (bright colored columns) and of 12 metagenomically detected strains (darker columns). Most outbreak samples cluster together due to almost identical profiles (right), with four samples (left) showing different profiles due to the presence of additional dominant E. coli strains overlying the target outbreak strain. (b) Functional analysis of outbreak-specific gene-families (Fisher exact test) confirmed that the outbreak strain is a combination of a EAEC pathogen (pAA plasmid) with acquired Shiga toxin and antibiotic resistance genes, complemented with a set of enriched virulence-related functions and pathway modules.
Large-scale population genomics study of E. rectale and A. muciniphila. Based on 1830 metagenomic samples from 8 cohorts, PanPhlAn reveals the subspecies structure even when only few species reference genomes are available. (a) Based on only one reference genome, E. rectale strains can be resolved into three geographically distinct clades. Clade A is related to samples of the two Chinese cohorts (bright and dark green dots). (b) Based on two available reference genomes, PanPhlAn shows a clear cluster structure of A. muciniphila strains, suggesting that the species can be distinguished into six functionally distinct clades A-F.
PanPhlAn is a project of the Computational Metagenomics Lab at CIBIO, University of Trento, Italy.
- PanPhlAn 3.0
- PanPhlAn 1.3