-
Notifications
You must be signed in to change notification settings - Fork 0
TechRequirements
Authors: Dave Walton and Keith Sheppard
This document describes the technical requirements for the first phase of the CGD Drake Genetics software project.
A mockup of what the entry point into the application might look like is below:

It is important to design the account management such that no direct technical support is required for a classroom to start using the DrakeGenetics web app after the server is up and running (though it will require some amount of system administration support to create a local instance of this web application). This is also intended to be a highly reusable component so an effort should be made to keep the software as modular as possible.
Any person with network access to the DrakeGenetics web application will be able to create a user account. Users will be uniquely identified by their email address which will be used to confirm the user's identity. All users will be able to:
- reset thier password via thier email address
- associate a name with thier account
- create a Laboratory
There will typcially be a one-to-one correspondance between classrooms using our application and Laboratories, though this relationship will not be enforced. Almost all communication between user accounts will occur within the context of a common Laboratory. The only exception to this being Laboratory invitations which clearly must be sent to users outside of the Laboratory.
When a Laboratory is created it will be initiallized with the following roles:
- Scientist (ie Student)
- Advisor (ie Teacher)
The user Account which created the Laboratory will be automatically assigned to the Advisor Role. Each item in the following activity list is followed by the role(s) which are authorized to perform that activity. All Advisor and Scientist permissions exist only in the context of the Laboratory where those roles are held:
- Laboratory creation: any user
- Laboratory account invitation/removal: Advisor
- Role assignment: Advisor
- Generate progress and usage reports: Advisor
- Read from notebook: Advisor, Scientist
- Write to own Laboratory notebook: Advisor, Scientist
- Read others' Laboratory notebook: Advisor, Scientist
- Comment on Laboratory notebook: Advisor, Scientist
- Perform Lab Experiments (TODO refer to a requirements section explaining which experiments are available): Scientist
- View Lab Experiment Results: Advisor, Scientist
- TODO: which permissions are missing
There will be no capability to create new roles or modify role permissions though this is something that we may consider adding in a future version.
Detailed Design information is available for Admin Module.
Each member of a Laboratory will have thier own Laboratory notebook.
Notebooks will be similar to blogs. Users with sufficient permissions will be able to read and comment on existing entries, as well as create new entries. Each entry will display an associated author and timestamp. Users must also have a convenient way to enter links which can be "permalinks" to experiment results, links to other notebook entries or external URLs.
- Rich text/HTML editor
- RSS feeds for notebook entries and comments
- Have a view that shows entries for all Laboratory members or just entries for a specific Laboratory member
Below is a mockup with an example of how the notebook might appear in a page. In this example, the notebook slides out from the side of the page. It would be available from any page in the interface.

Multiple choice quizes will be authored using the Aiken format. The name of the quiz will be derived from the filename using the following simple algorithm. The filename "the-basics-of-meiosis.aiken" will be used as an example:
- first if there is a file extension it will be removed: "the-basics-of-meiosis.aiken" => "the-basics-of-meiosis"
- all dashes "-" or underscores "_" will be replaced by spaces: "the-basics-of-meiosis" => "the basics of meiosis"
Minimum requirements: at a minimum system administrators must be able to upload tests using an administrative interface. This can be a web interface, or simply a command-line interface.
Optional: Allow Advisors to upload custom tests for thier virtual Laboratories.
Advisors will be able to see test results for all Scientists in thier Laboratories. There will be a main test overview page where quiz results will be summarized in a table where the rows correspond to Scientists and the columns correspond to tests. The table cells will also be linked to test detail pages where the Advisors will be able to see the answers given by a student for a particular test. Scientists will be able to view thier own test results but will not be able to see anyone elses test results.
Track time on task and student workflow (i.e. the "path" they follow while using the application). TODO: we need more specifics regarding what constitutes a task and what a useful report will contain before we can fill out the technical requirements for this section.
Most of the requirements up until this point have been fairly generic. The requirements in this section are much more specific to the Drake Genetics application.
Below is a mockup of what the Breeding module in the interface may look like:
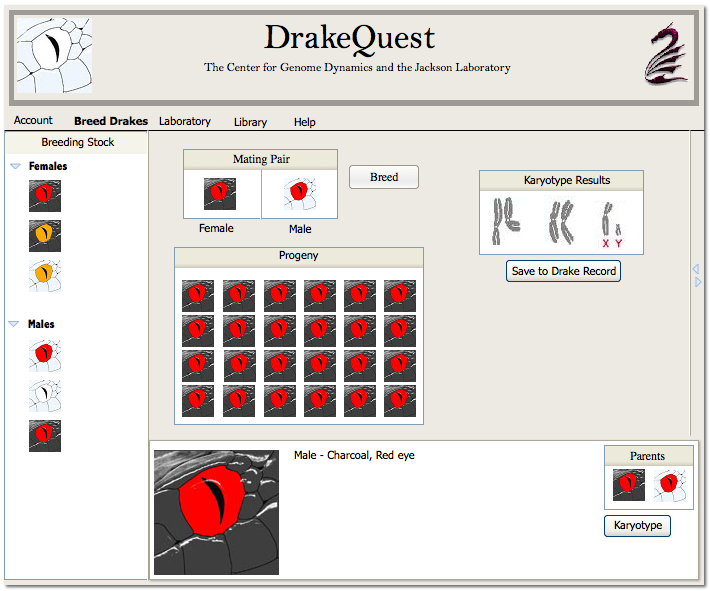
When a pair of drakes is bred, the mieosis engine must be used to simulate sexual reproduction for each of the fertilized eggs produced. So, for each egg we will use the meiosis engine to randomly and independently generate gamete DNA for the mothers egg and the fathers sperm. We will then combine these two sources of gamete DNA in order to create the DNA for a fertilized egg.
TODO: determine how breeding pair is selected. From fixed initial stock? from random stock? One of the concepts is that students will be able to breed and exchange dragons but I think we'll have to consider the exchange of dragon stocks to be an optional requirement for the moment.
The breeding pair is selected by the student using one of several methods. A breeding pair may be supplied to the student when they undertake a Quest. A student may purchase drakes in the market, or take them from his or her stable. Drakes to be bred are imported into the Breeding Pen. One male and one female must be brought into the breeding pen before anything else can happen. The female occupies the position on the left (always) in the display, and the male occupies the position on the right. An attempt to import two males or two females will generate an error message. If the parentage of either drake is known, the parental phenotypes are displayed.
The parentage will be known if the student has bred the drake or if the drake is from an inbred strain. For some drakes purchased in the market or supplied when Quests are undertaken, the parentage will be unknown.
There is some probability that the chromosomes will experience nondisjunction during meiosis. Sex chromosomes will experience nondisjunction with a probability of 0.01 while autosomes will experience nondisjunction with a probability of 0.002. Once you have determined that a particular parental chromosome has experienced nondisjunction there are a couple of special considerations you must make:
- in the case of nondisjunction you must give equal (0.5) probability to the case possible outcomes: that the fertilizing gamete has either 0 or 2 copies of the affected chromosome
- TODO fill in details about how nondisjunction affects DNA at the centromere. Is meiosis affected in other ways?
Let me talk you through the meiosis engine in a slightly more elaborated form than what we first discussed.
-
Check for fertility. If the female is X0 or has any other form of genetically determined female sterility, no eggs will be produced. If the male is XXY or has any other form of genetically determined male sterility, the female will lay a clutch of unfertilized eggs. The chromosome content is irrelevant in these cases.
-
Replication. In fertile crosses, for each chromosome, replicate to form two identical sister chromatids (that's what they are called). They are exact copies.
-
Determine position of crossovers. There is one exception; there is no crossing over between the X and Y chromosome in males. First, randomly select the 10 cM segments that will experience crossover events. Give every 10 cM segment of the chromosome an independent probability of 0.1 of experiencing a crossover. This will result in you having between 0 and num_10cM_segments crossover events.
If the number of crossover events is greater than three then randomly drop all but three of the crossover events. For each remaining event we will determine the exact location of the crossover using a uniform random distribution from the start to the end of the 10 cM segment.
-
Determine the chromatids involved in each crossover and resolve the crossovers. Imagine that one chromosome has replicated to make chromatids 1 and 2. The other chromosome has replicated to make chromatids 3 and 4. For each crossover, select at random either chromatid 1 or chromatid 2. The partner for the crossover will be either chromatid 3 or chromatid 4, selected at random.
-
Segregate chromosomes into gametes. This is the point at which we determine if segregation occurs normally (p = 0.99 for sex chromosomes, p = 0.998 for autosomes) or whether nondisjunction occurs.
In case of nondisjunction, half of the time the gametes will get no chromatids for that chromosome. The other half of the time the gamete will get two chromatids. In the case of a gamete with two chromatids, the first chromatid will be chosen at random. The second chromatid will contain one of the two nonsister centromeres. For example, in the simplest case where there are no crossovers, if chromatid 1 is chosen, either chromatid 3 or chromatid 4 will be chosen as the other chromatid. Another simple case is the sex chromosome pair in males. All gametes with sex chromosome nondisjunction in males will have one X and one Y chromosome.
- Determine whether the zygote is viable or will have arrested development (from phenotype lookup table, which includes aneuploids).
We will employ the following algorithm to determine crossover locations:
- First randomly select the 10cM segments that will experience crossover events. Give every 10cM segment of the chromosome an independent probability of 0.1 of experiencing a crossover. This will result in you having between 0 and num_10cM_segments crossover events.
- If the number of crossover events is greater than three then randomly drop all but three of the crossover events. TODO: confirm that this is the intent
- for each remaining event we will determine the exact location of the crossover using a uniform random distribution from the start to the end of the 10cM segment
Phenotype will be determined by a combination of karyotype (in the case of nondisjunction), genotype and diet (NOTE: diet will be optional for the initial phase of development).
This module is based on a Thesis written by Abby Todd in 2008 for fulfillment of the requirements for a PHD at UNC Chapel Hill in the Department of Mathematics. Parts of this text below is drawn directly from the thesis.
The Metabolism Module, will allow a Scientist/Student to select a Drake for their experiment.
The basic experiment will allow them to feed their Drake a normal diet.
Once fed, the scientists will be able to monitor the Drake over a time period, measuring various aspects of the specimen’s metabolic system. In the simplest scenario, the specimen’s blood glucose will be measured.
There is an assumption that all experiments start at a “fed state” and that we are measuring the affects of fasting over a time period. A more advanced experiment could take the Drake to a fasted or near fasted state, and then feed the drake again, and monitor the specimen over time as the system transitions back from a fasted to fed state. In the initial release of this module we will just worry about what happens to the drake as it transitions from a fed state to a fasted state.
Another more advanced experiment could be to vary the diet. The 3 main components of the diet include protein, carbohydrates and fats. In this advanced experiment the scientist could increase or decrease the amounts of any of these three and then repeat the process of monitoring the metabolic system to determine the differences in the outcome as the specimen transitions from fed to fasted state.
The mathematical model on which our metabolism simulator will be built has models for blood (the transport system for metabolites) and three tissues:
- Liver (control center for the metabolic system)
- Fat (for regulating temperature, insulating body organs and storage of important excess metabolic fuels)
- Muscle (in this model it behaves primarily as a consumer of glucose)
The main goal of this module, is to allow the scientist to explore the dynamic shift in concentration levels for the main metabolic fuels, when the system is adapting from a fed to fasted state. The scientist should be able to measure relative change in:
- blood glucose
- liver glycogen
- plasma free fatty acids
- blood ketone bodies
- plasma insulin
- plasma glucagon (NOTE DOW: Need to confirm that all of these can be obtained form the UNC Matlab code)
Below is an example of what the interface to the Metabolism Module could look like.

QUESTION DOW: Should scientist be able to control reduced or full regulatory circuit? What does this mean? How would they control this?
The library is intended as a location where a student/scientist can go to learn about Drakes, breeding, genetics, protocols, metabolism, etc... A simple interface will be provided (seen below), that will allow the student to browse through the available journal articles in the Library and read them.

The logic that supports this will be fairly simple, and will assume that the contents will be stored in a directory structure, where each layer in the directory structure represents a heading in the hierarchical tree panel on the left (under "Publications"). The directory structure should not be more than 3 layers deep, and at the last layer, there should be a set of files, where each file represents a journal article that can be displayed in the right hand panel. The name of the file will be used as the name of the item in the browsing panel on the left (DOW: Do we want a definition file for a human readable article name?). The panel on the right will have tabs so that the user can have multiple articles open at a time.
From any page in the interface the user should be able to access help. The help has a structure very similar to the Library. The differences being that the Help will come up as a free dialog box, which can be referenced while still working in the interface, and the document supplied are help pages not publications. Below is an example of how the help dialog might look if brought up from the breeding page.
