-
Notifications
You must be signed in to change notification settings - Fork 2
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Showing
1 changed file
with
44 additions
and
0 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1 +1,45 @@ | ||
| # Project-Arrhythmia | ||
|
|
||
|  | ||
|
|
||
| ### Introduction | ||
|
|
||
| The Dataset used in this project is available on the UCI machine learning Repository. | ||
| It can be found at: [https://archive.ics.uci.edu/ml/datasets/Arrhythmia](https://archive.ics.uci.edu/ml/datasets/Arrhythmia). | ||
| * It consists of 452 different examples spread over 16 classes. Of the 452 examples,245 are of "normal" people. | ||
| * We also have 12 different types of arrhythmias. Among all these types of arrhythmias, the most representative are the "coronary artery disease" and "Rjgbt boundle branch block". | ||
| * We have 279 features, which include age, sex, weight, height of patients and other related information. | ||
| * We explicitly observe that the number of features is relatively high compared to the number of examples we are available. | ||
| * Our goal was to predict if a person is suffering from arrhythmia or not, and if yes, classify it in to one of 12 available groups. | ||
|
|
||
| ### Directory Structure | ||
|
|
||
| * [1- Reports and presentations](https://github.com/shsarv/Project-Arrhythmia/tree/master/1-%20Reports%20and%20presentations) - Contains project report, presentation and papers. | ||
| * [Data Preprocessing Notebook](https://github.com/shsarv/Project-Arrhythmia/blob/master/Preprocessing%20and%20EDA/Data%20preprocessing.ipynb) | ||
| * [Exploratory Data Analysis Notebook ](https://github.com/shsarv/Project-Arrhythmia/blob/master/Preprocessing%20and%20EDA/EDA.ipynb) | ||
| * [Final Notebook with pca](https://github.com/shsarv/Project-Arrhythmia/blob/master/final%20with%20pca.ipynb) | ||
| * [Oversampled notebook at ](https://github.com/shsarv/Project-Arrhythmia/blob/master/Model/oversampled%20and%20pca.ipynb) | ||
|
|
||
|
|
||
|
|
||
| ### Algorithms Used | ||
| 1. KNN Classifier | ||
| 2. Logestic Regression | ||
| 3. Decision Tree Classifier | ||
| 4. Linear SVC | ||
| 5. Kernelized SVC | ||
| 6. Random Forest Classifier | ||
| 7. Principal Component analysis (PCA) | ||
|
|
||
| ### Result | ||
|
|
||
| 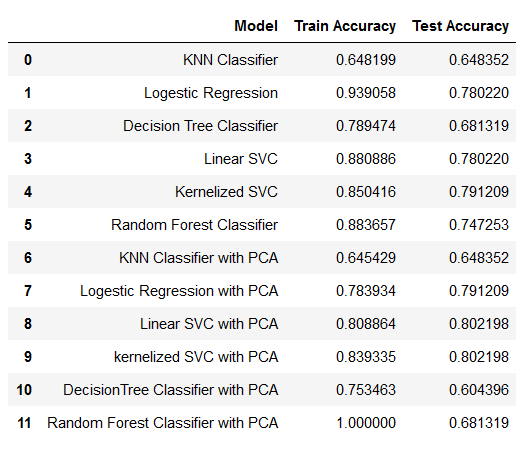 | ||
|
|
||
|
|
||
| ### Conclusion | ||
|
|
||
| The models started performing better after we applied PCA on the resampled data. The reason behind this is, PCA reduces the complexity of the data. It creates components based on giving importance to variables with large variance and also the components which it creates are non collinear in nature which means it takes care of collinearity in large data set. PCA also improves the overall execution time and quality of the models and it is very beneficial when we are working with huge amount of variables. | ||
|
|
||
| **The Best model in term of recall score is Kernalized SVM with PCA having accuracy of 80.21%.** | ||
|
|
||
|
|